My Projects
I have been involved in research since 2011 and have had the oppurtunity to work on a variety of projects.
As an undergraduate I did research in the lab of Catherine Putonti at Loyola University Chicago. Here I primarily studied phage-host interactions via the CRISPR system. I developed a tool for use in the lab to match CRISPR spacer seqeunces from microbial genomes to phage genomes and generate an interaction network.
As a graudate student I studied in Catherine Lozupone's lab at University of Colorado Anschutz Medical Campus where I worked on developing methods 1) for correlation network analysis of compositional data such as 16S rRNA amplicon sequencing and untarged LC/MS metabolomics and 2) connecting the genomic content of microbes to compounds they could potential produce/consume. In addition to these bioinformatics software development projects I also applied these methods, and others, to researching the effect of HIV on the human gut microbiome.
I am currently a postdoc in Kelly Wrighton's lab at Colorado State University. I've had the oppurtunity to work on a wide variety of projects in the Wrighton lab. DRAM is my primary software development project. It is a tool that will annotate bacterial genomes/metagenomes and tell you what functions they are potentially capable of. I built all of the software while the metabolism and other genome function was curated by a team of experts led by my co-first author Mikayla Borton. My primary biologly focused work is in studying the effects Salmonella infection on the gut microbial community in mice. This work invovles collaboration with the Ahmer and Wysocki labs at Ohio State University and I analyze the sequencing based 'omics and lead the integration between mass spec based 'omics and the sequencing based 'omics. Additionally I've gotten to help out with other projects doing statistical analysis as well as bioinformatics pipeline development in environments including the human gut, global river systems, wetland sediments and permafrost soils.
Here is some more detail on some of the projects I'm most proud of.
DRAM: Annotating and summarizing functional content of microbial genomes/metagenomes
DRAM more than a genome annotation tool. While it performs this function, and does very well at it in comparison to similar software, what really sets DRAM apart is the ability to summarize the functions that are present in a genome/metagenome in an easy to understand way. It distills this information to generate three levels for users to examine. The raw contains all annotations for all genes from all databases. The distillate gives an overview of all the functional genes which are present across all genomes. The product gives an interative heatmap allowing the user to explore a high level overview of the functions a acollection of genomes are capable of. You can check out what the product looks like by clicking here.
In order to make DRAM available to a wider audience we have also developed a series of KBase apps. KBase is a cyberinfrastructure developed by the Department of Energy with the goal of making 'omics analysis available to everyone. You can read more about the project at www.kbase.us. The DRAM KBase apps allow users to access DRAM through a web interface and without needing access to powerful servers. The DRAM KBase apps also allow for integration into the larger KBase system of tools including using DRAM annotations in flux balance analysis simulations.
DRAM recieved over 20 citations in the first year it was published. You can check it out in Nucleic Acids Research. We also had the opportunity to present DRAM as part of the Ohio State University Center of Microbiome Science webinar series which you can watch online here. DRAM development continues on it's github page where there is also a wiki full of helpful documentation.
2020 Multiscale Microbial Dynamics Modeling Summer School
In 2020 I had the opportunity to act as an instructor for the 2020 DOE Environmental Molecular Sciences Laboratory summer school. This is a program put on each summer by EMSL with a different theme every year. The 2020 edition focused on microbial dynamics modeling. Ordinarily this program is run in person but this was not possible due to the COVID-19 pandemic. As a result it was taught online and the general lectures were opened up to anyone who wanted to attend while the small group projects were run online. I taught a module about genome annotation and how DRAM can be used to annotate gene function. The talk I gave is available on youtube. Additionally I build the KBase narratives that were used by the students and led a student group during the event. As a cap to this work I presented a talk at the 2020 AGU fall meeting about the discoveries we were able to make in collaborations with the students who participated.
SCNIC: Building and analyzing correlation networks made from compositonal data
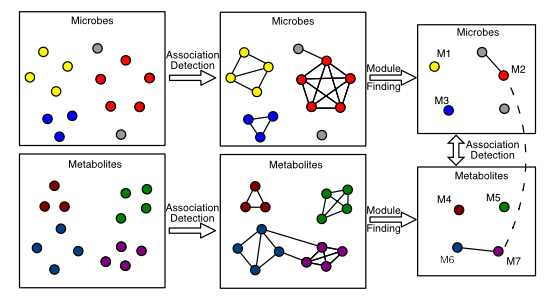
An important part of my PhD thesis was developing methods for correlation network analysis with compositional data. Compositional data, such as 16S amplicon sequencing or untargeted LC/MS metabolomics, needs to be treated differently than count data. Methods to take compositionality into account have been developed but are not used in many of the most popular tools for building and finding modules in correlation networks. To address these issues I developed a tool called SCNIC. SCNIC builds correlation networks from compositional data, as well as other data types, and then finds modules of correlated observations, via user selected methods. The SCNIC manuscript is available as a preprint. To make SCNIC more useful for the 16S rRNA amplicon sequencing community SCNIC is also available as a QIIME2 plugin.
AMON: Determining potential metabolite origin from microbial genomes
During my PhD I also sought to make it easier for users to explain the relationships between bacteria genomes and the metabolites they can produce and consume. To do this I build AMON. AMON takes as input lists of KEGG KO identifiers and then generates a list of metabolites that could be produced and consumed by those KOs. This is done by querying the KEGG API (or the user providing KEGG flat files) and then parsing them to find the inputs and outputs of the reactions associated with a set of KEGG KO IDs. The AMON source code is available on github. The AMON manuscript was published in BMC Bioinformatics.